![]() CONTINUE
CONTINUE
A new innovative technique developed by a scientific team at the Center for Genomic Regulation (CRG) in Barcelona has discovered the existence of a multitude of 'remote controls' that control the function of proteins and that can be used as targets to achieve more effective drugs and efficient in various pathologies such as dementia, cancer and infectious infections.
These 'remote controls' are scientifically known as allosteric sites. These are remote controls that are distant from the site of action of the protein, but have the capacity to regulate or modulate it", Júlia Domingo, first co-author of the study, which is published this Wednesday in the journal "Nature", explained to ABC. She and she adds a simile: "It's as if with that remote control you could turn the light bulb on and off or regulate the intensity of the light."
In this case where it intends to block or regulate the activity of proteins that maintain their altered function in confinement. For example, in the case of cancer, the proteins that have acquired a mutation have their functionality altered, they do so abnormally and the cell grows unusually. In many cases, there are no drugs that can modulate or block this abnormal activity or, if there are, they are not specific and are also released from other proteins that function normally.
Traditionally, drug hunters have designed treatments that target the active site of a protein, whose small region produces chemical reactions where targets bind. The drawback of these drugs, known as orthosteric drugs, is that the active sites of many proteins are very similar and the drugs have bound and inhibited many different proteins at the same time, even those that function normally and are not interesting to touch, which can cause side effects.
“There he entered the concept of allosteria and the potential it has to design drugs. The interesting thing about allosteric sites is that they are super specific for each protein. If these allosteric sites find part of the protein surface where the drug can land, it will be extremely specific for that protein. We will be able to aspire to more effective medicines”, points out the researcher.
“Not only do we find that these therapeutic sites are abundant, but there is evidence that they can be manipulated in many different ways. Instead of just turning them on and off, we can modulate their activity like a thermostat. From an engineering point of view, it is as if we have struck gold, because it gives us a lot of room to design 'smart drugs' that go to the bad and skip the good", explains André Faure, postdoctoral researcher at the CRG and first co - author of the article.
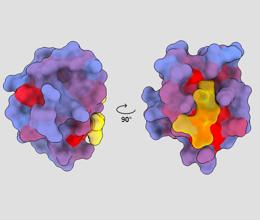 three-dimensional image showing the human protein PSD95-PDZ3 from different points of view. A molecule is shown binding to the active site in yellow. Blue to red color gradient indicates possible allosteric sites – André Faure/ChimeraX
three-dimensional image showing the human protein PSD95-PDZ3 from different points of view. A molecule is shown binding to the active site in yellow. Blue to red color gradient indicates possible allosteric sites – André Faure/ChimeraX
For this discovery, the team has used a method that allows them to take a protein and a systemic form and a global encounter with all the sites. To do this, they have chosen two very abundant proteins in our human proteome. “50% of the protein surface has allosteric potential. Our method makes it possible to make an atlas of allosteric sites, which would make the process of searching for effective drugs much more efficient”, assures Júlia Domingo.
The study authors developed a technique called double-depth PCA (ddPCA), which they describe as a "brute force experiment." "We purposely break things in thousands of different ways to form a complete picture of how something works," explains ICREA Research Professor Ben Lehner, Coordinator of the Systems Biology program at CRG and an author of the study. “It's like if you suspect a spark plug is bad, but instead of just checking that, the mechanic will take the whole car apart and check all the parts one by one. By analyzing ten thousand things at once, we identify all the pieces that are really important.”
Next, we use artificial intelligence algorithms to interpret the lab results.
One of the great advantages of the method, in addition to simplifying the process necessary to find allosteric sites, is that it is an affordable and accessible technique for any research laboratory in the world. “It just requires access to basic molecular biology reagents, access to a DNA sequencer and a computer. With these three components, any laboratory in 2-3 months, with a small budget, can carry out this experiment on the protein of interest that they want”, assures Júlia Domingo. The researchers' hope is that our scientists will use the technique to quickly and comprehensively map the allosteric sites of human proteins one by one. “If we have enough data maybe one day we can go one step further and predict from protein sequence to function. Use these data to guide them as better therapies to predict if a certain change in a protein is going to degenerate into a disease”, concluded the researcher.
